4 Dvosmjerna analiza varijance
Nekoliko uvodnih riječi…
Nekakvi tekst pa nabrajanje
- opcija 1 tekst
- opcija 2
- opcija 3
Neka fusnota2
4.1 Primjer (bez interakcije)
Primjer u kojemu pretpostavljamo da nema interakcije između dvije faktorske varijable.
ToothGrowth$dose <- factor(ToothGrowth$dose)
a2_model <- aov(len ~ supp + dose, data = ToothGrowth)
print(summary(a2_model), digits = 7)## Df Sum Sq Mean Sq F value Pr(>F)
## supp 1 205.350 205.3500 14.01664 0.00042928 ***
## dose 2 2426.434 1213.2172 82.81093 < 2.22e-16 ***
## Residuals 56 820.425 14.6504
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Ručni izračun
tablica_ab <- ToothGrowth %>%
group_by(supp) %>% mutate(Ya = mean(len)) %>% ungroup() %>%
group_by(dose) %>% mutate(Yb = mean(len)) %>% ungroup()
Ybar <- mean(ToothGrowth$len)
SSA <- tablica_ab %>% summarize(SSA = sum((Ya - Ybar)^2)) %>% pull(SSA)
SSB <- tablica_ab %>% summarize(SSB = sum((Yb - Ybar)^2)) %>% pull(SSB)
SST <- tablica_ab %>% summarize(SST = sum((len - Ybar)^2)) %>% pull(SST)
SSR <- SST - SSA - SSB
dfA <- nlevels(ToothGrowth$supp) - 1
dfB <- nlevels(ToothGrowth$dose) - 1
dfR <- nrow(ToothGrowth) - 1 - dfA - dfB
MSA <- SSA / dfA
MSB <- SSB / dfB
MSR <- SSR / dfR
FA <- MSA / MSR
FB <- MSB / MSR
pA <- pf(FA, df1=dfA, df2=dfR, lower.tail=FALSE)
pB <- pf(FB, df1=dfB, df2=dfR, lower.tail=FALSE)
list(SSA = SSA, SSB = SSB, SSR = SSR, dfA = dfA, dfB = dfB, dfR = dfR,
MSA = MSA, MSB = MSB, MSR = MSR, FA = FA, FB = FB,
pA = pA, pB = pB)## $SSA
## [1] 205.35
##
## $SSB
## [1] 2426.434
##
## $SSR
## [1] 820.425
##
## $dfA
## [1] 1
##
## $dfB
## [1] 2
##
## $dfR
## [1] 56
##
## $MSA
## [1] 205.35
##
## $MSB
## [1] 1213.217
##
## $MSR
## [1] 14.65045
##
## $FA
## [1] 14.01664
##
## $FB
## [1] 82.81093
##
## $pA
## [1] 0.0004292793
##
## $pB
## [1] 1.871163e-17Effect size
etaSquared(x = a2_model)## eta.sq eta.sq.part
## supp 0.05948365 0.2001901
## dose 0.70286419 0.7473174Isti rezultat možemo dobiti i ručnim izračunom.
list(eta.sq.SUPP = SSA / SST, eta.sq.part.SUPP = SSA / (SSA + SSR),
ets.sq.DOSE = SSB / SST, eta.sq.part.DOSE = SSB / (SSB + SSR))## $eta.sq.SUPP
## [1] 0.05948365
##
## $eta.sq.part.SUPP
## [1] 0.2001901
##
## $ets.sq.DOSE
## [1] 0.7028642
##
## $eta.sq.part.DOSE
## [1] 0.74731744.2 Primjer (s interakcijom)
Primjer u kojemu pretpostavljamo da ima interakcije između dvije faktorske varijable.
ToothGrowth %>%
ggplot(aes(x = supp, y=len, color = dose, group = dose)) +
stat_summary(fun.y = mean, geom = "point") +
stat_summary(fun.y = mean, geom = "line")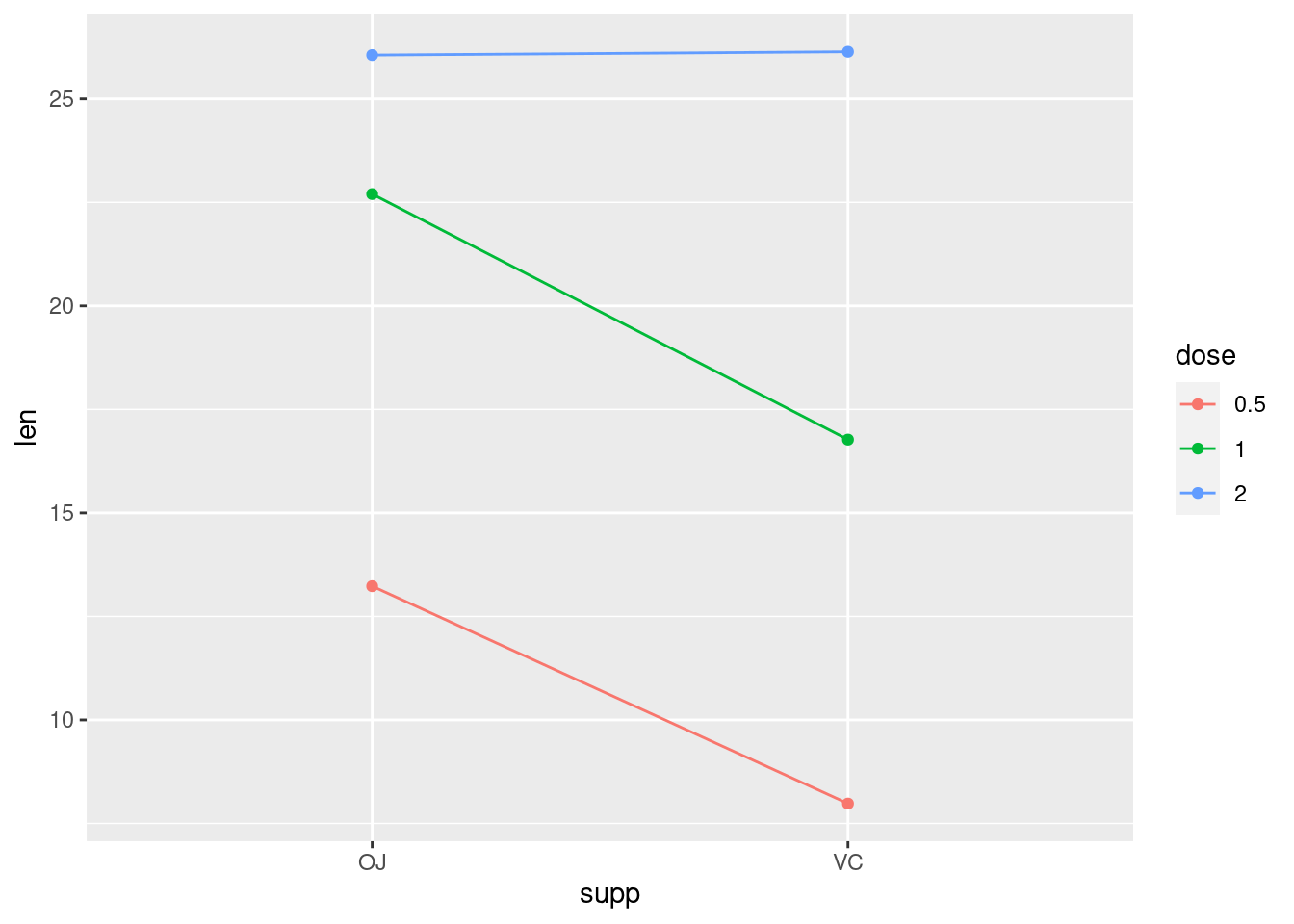
ToothGrowth %>%
ggplot(aes(x = dose, y=len, color = supp, group = supp)) +
stat_summary(fun.y = mean, geom = "point") +
stat_summary(fun.y = mean, geom = "line")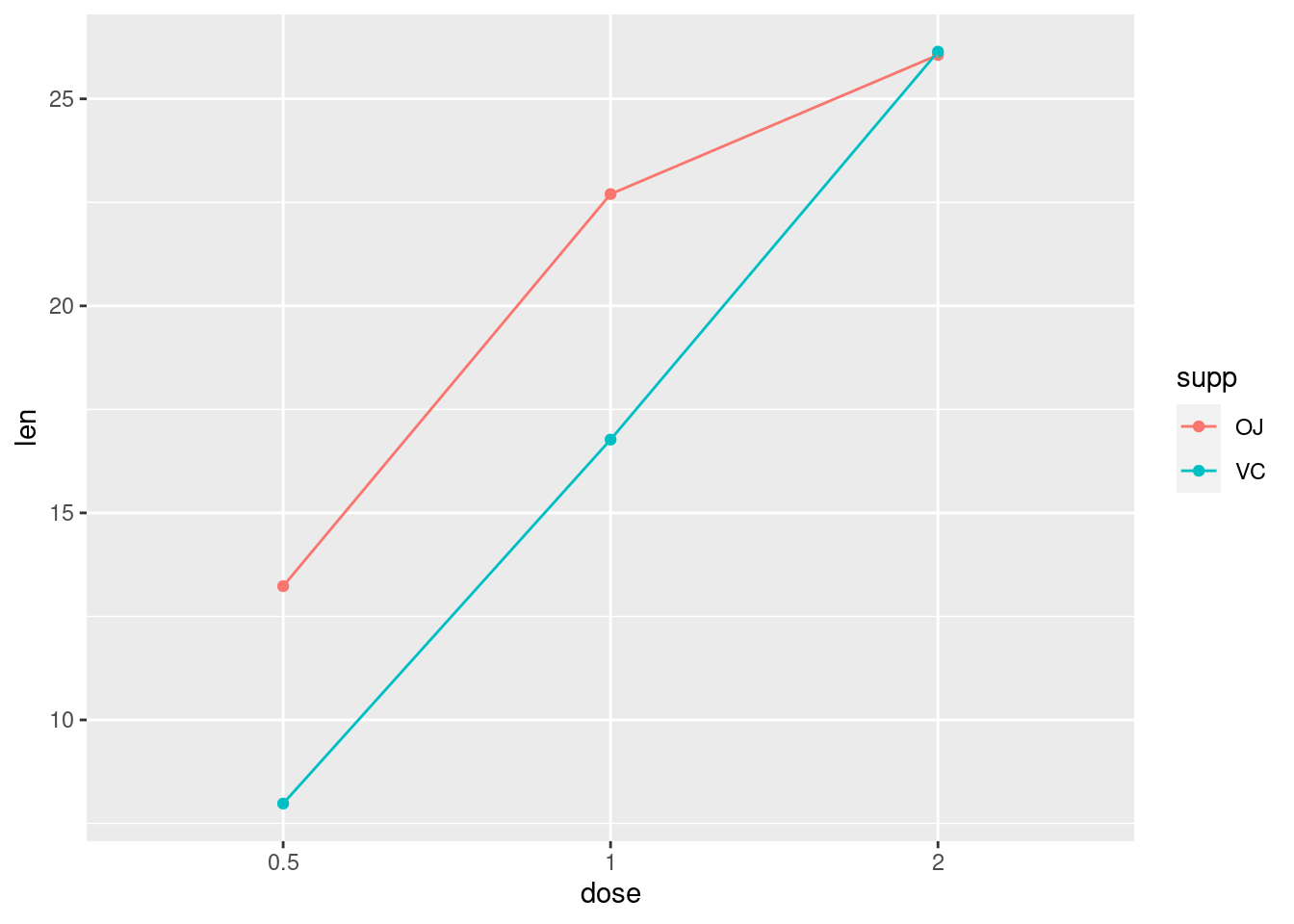
a2_inter <- aov(len ~ supp * dose, data = ToothGrowth)
print(summary(a2_inter), digits = 7)## Df Sum Sq Mean Sq F value Pr(>F)
## supp 1 205.350 205.3500 15.57198 0.00023118 ***
## dose 2 2426.434 1213.2172 91.99996 < 2.22e-16 ***
## supp:dose 2 108.319 54.1595 4.10699 0.02186027 *
## Residuals 54 712.106 13.1871
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Ručni izračun
tablica_ab_inter <- ToothGrowth %>% group_by(supp, dose) %>%
mutate(Yab = mean(len)) %>% ungroup %>%
group_by(supp) %>% mutate(Ya = mean(len)) %>% ungroup() %>%
group_by(dose) %>% mutate(Yb = mean(len)) %>% ungroup()
Ybar <- mean(ToothGrowth$len)
SSA <- tablica_ab_inter %>% summarize(SSA = sum((Ya - Ybar)^2)) %>% pull(SSA)
SSB <- tablica_ab_inter %>% summarize(SSB = sum((Yb - Ybar)^2)) %>% pull(SSB)
SSAB <- tablica_ab_inter %>% summarize(SSAB = sum((Yab - Ya - Yb + Ybar)^2)) %>%
pull(SSAB)
SST <- tablica_ab_inter %>% summarize(SST = sum((len - Ybar)^2)) %>% pull(SST)
SSR <- SST - SSA - SSB - SSAB
dfA <- nlevels(ToothGrowth$supp) - 1
dfB <- nlevels(ToothGrowth$dose) - 1
dfAB <- dfA * dfB
dfR <- nrow(ToothGrowth) - (dfA + 1) * (dfB + 1)
MSA <- SSA / dfA
MSB <- SSB / dfB
MSAB <- SSAB / dfAB
MSR <- SSR / dfR
FA <- MSA / MSR
FB <- MSB / MSR
FAB <- MSAB / MSR
pA <- pf(FA, df1=dfA, df2=dfR, lower.tail=FALSE)
pB <- pf(FB, df1=dfB, df2=dfR, lower.tail=FALSE)
pAB <- pf(FAB, df1=dfB, df2=dfR, lower.tail=FALSE)
list(SSA = SSA, SSB = SSB, SSAB = SSAB, SSR = SSR,
dfA = dfA, dfB = dfB, dfAB = dfAB, dfR = dfR,
MSA = MSA, MSB = MSB, MSAB = MSAB, MSR = MSR,
FA = FA, FB = FB, FAB = FAB,
pA = pA, pB = pB, pAB = pAB)## $SSA
## [1] 205.35
##
## $SSB
## [1] 2426.434
##
## $SSAB
## [1] 108.319
##
## $SSR
## [1] 712.106
##
## $dfA
## [1] 1
##
## $dfB
## [1] 2
##
## $dfAB
## [1] 2
##
## $dfR
## [1] 54
##
## $MSA
## [1] 205.35
##
## $MSB
## [1] 1213.217
##
## $MSAB
## [1] 54.1595
##
## $MSR
## [1] 13.18715
##
## $FA
## [1] 15.57198
##
## $FB
## [1] 91.99996
##
## $FAB
## [1] 4.106991
##
## $pA
## [1] 0.0002311828
##
## $pB
## [1] 4.046291e-18
##
## $pAB
## [1] 0.02186027Effect size
etaSquared(x = a2_inter)## eta.sq eta.sq.part
## supp 0.05948365 0.2238254
## dose 0.70286419 0.7731092
## supp:dose 0.03137672 0.1320279Isti rezultat možemo dobiti i ručnim izračunom.
list(eta.sq.SUPP = SSA / SST, eta.sq.part.SUPP = SSA / (SSA + SSR),
ets.sq.DOSE = SSB / SST, eta.sq.part.DOSE = SSB / (SSB + SSR),
eta.sq.SUPP.DOSE = SSAB / SST, eta.sq.part.SUPP.DOSE = SSAB / (SSAB + SSR))## $eta.sq.SUPP
## [1] 0.05948365
##
## $eta.sq.part.SUPP
## [1] 0.2238254
##
## $ets.sq.DOSE
## [1] 0.7028642
##
## $eta.sq.part.DOSE
## [1] 0.7731092
##
## $eta.sq.SUPP.DOSE
## [1] 0.03137672
##
## $eta.sq.part.SUPP.DOSE
## [1] 0.13202794.3 Levene test
This function expects that you have a saturated model (i.e., included all of the relevant terms), because the test is primarily concerned with the within-group variance, and it doesn’t really make a lot of sense to calculate this any way other than with respect to the full model.
leveneTest(a2_model, center = mean)## Error in leveneTest.formula(f, data = m, ...): Model must be completely crossed formula only.leveneTest(a2_inter, center = mean)## Levene's Test for Homogeneity of Variance (center = mean)
## Df F value Pr(>F)
## group 5 1.9401 0.1027
## 544.4 Normalnost reziduala
Pogledajmo reziduale za model s interakcijom.
a2.residuals <- tibble(res = residuals(object = a2_inter))
a2.residuals## # A tibble: 60 × 1
## res
## <dbl>
## 1 -3.7800
## 2 3.5200
## 3 -0.68000
## 4 -2.1800
## 5 -1.5800
## 6 2.0200
## 7 3.2200
## 8 3.2200
## 9 -2.7800
## 10 -0.98000
## # … with 50 more rowsHistogram
ggplot(a2.residuals, aes(x=res)) + geom_histogram(binwidth=1, fill="pink", color="red")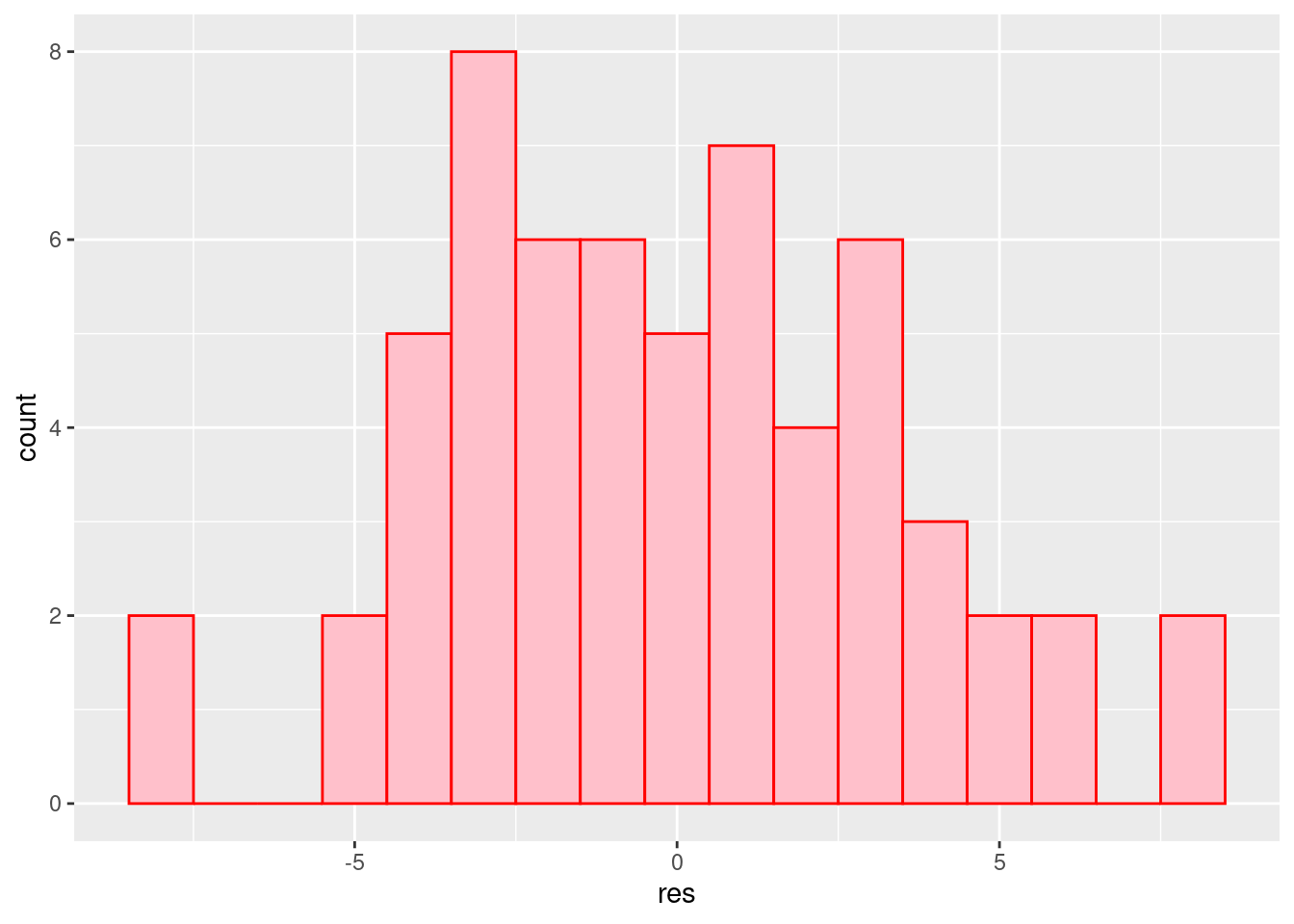
QQ-plot
ggplot(a2.residuals, aes(sample = res)) + stat_qq() + stat_qq_line()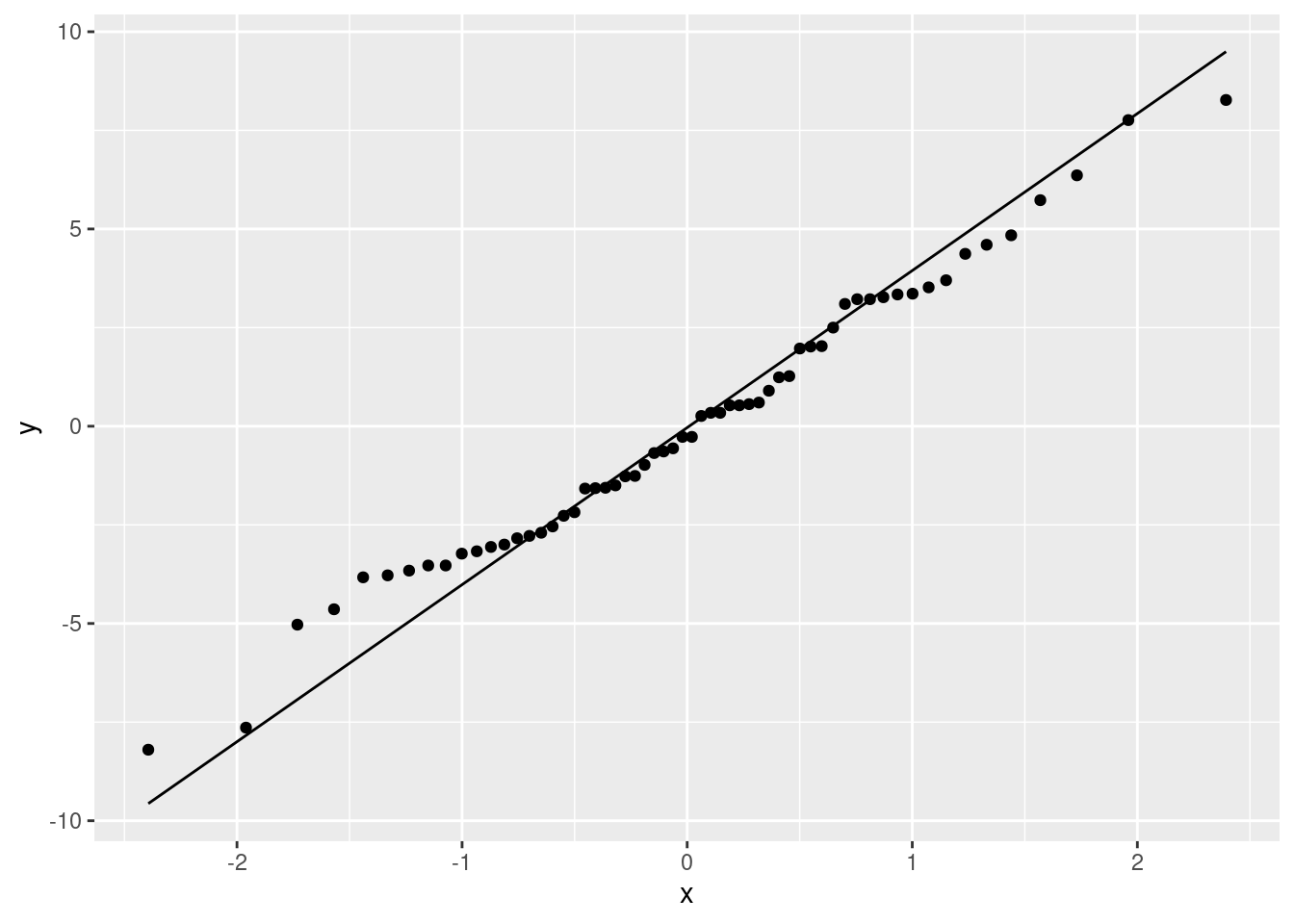
Shapiro-Wilk test za ispitivanje normalnosti
Početna hipoteza: “Populacija ima normalnu distribuciju.”
shapiro.test(a2.residuals$res)##
## Shapiro-Wilk normality test
##
## data: a2.residuals$res
## W = 0.98499, p-value = 0.66944.5 Biranje modela
Bolji model u ovom slučaju je model s uključenom interakcijom.
anova(a2_model, a2_inter)## Analysis of Variance Table
##
## Model 1: len ~ supp + dose
## Model 2: len ~ supp * dose
## Res.Df RSS Df Sum of Sq F Pr(>F)
## 1 56 820.43
## 2 54 712.11 2 108.32 4.107 0.02186 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Ručni izračun
- Stupnjevi slobode definirani su u smislu broja parametara koji se moraju procijeniti u modelu.
- F-test je uspoređivanje dva linearna modela.
- Prvi stupanj slobobde \(\mathrm{d}f_1\) u F-testu jednak je razlici broja parametara koje treba procijeniti u dva modela koji se uspoređuju.
- Drugi stupanj slobode \(\mathrm{d}f_2\) u F-testu odnosi se na stupnjeve slobode povezane s rezidualima. To je zapravo jednako razlici ukupnog broja opažanja (broj ispitanika) i broja parametara koje treba procijeniti.
U slučaju 2-anove, ukoliko faktorske varijable imaju redom \(a\) i \(b\) faktora, tada se svaka od njih može
redom reprezentirati s \(a-1\) i \(b-1\) binarnih varijabli. Za model bez interakcije treba procijenti ukupno
\((a-1)+(b-1)+1=a+b-1\) parametara (parametar uz svaku binarnu varijablu i slobodni član).
Za puni model sa svim interakcijama treba procijeniti ukupno \(ab\) parametara.
\[ab=((a-1)+1)((b-1)+1)=(a-1)+(b-1)+(a-1)(b-1)+1\]
Naime, uz svaku binarnu varijablu imamo \((a-1)+(b-1)\) parametara, uz sve moguće interakcije imamo
\((a-1)(b-1)\) parametara i konačno još jedan parametar za slobodni član.
a2_model_tidy <- tidy(a2_model)
a2_inter_tidy <- tidy(a2_inter)
a2_model_tidy## # A tibble: 3 × 6
## term df sumsq meansq statistic p.value
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 supp 1 205.35 205.35 14.017 4.2928e- 4
## 2 dose 2 2426.4 1213.2 82.811 1.8712e-17
## 3 Residuals 56 820.43 14.650 NA NAa2_inter_tidy## # A tibble: 4 × 6
## term df sumsq meansq statistic p.value
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 supp 1 205.35 205.35 15.572 2.3118e- 4
## 2 dose 2 2426.4 1213.2 92.000 4.0463e-18
## 3 supp:dose 2 108.32 54.160 4.1070 2.1860e- 2
## 4 Residuals 54 712.11 13.187 NA NAa <- nlevels(ToothGrowth$supp)
b <- nlevels(ToothGrowth$dose)
SSR.null <- a2_model_tidy %>% pull(sumsq) %>% last()
SSR.full <- a2_inter_tidy %>% pull(sumsq) %>% last()
SSR.diff <- SSR.null - SSR.full
df1 <- a * b - (a + b - 1)
df2 <- nrow(ToothGrowth) - a * b
MS.diff <- SSR.diff / df1
MS.res <- SSR.full / df2
Fv <- MS.diff / MS.res
pvalue <- pf(Fv, df1, df2, lower.tail = FALSE)
list(SSR.null = SSR.null, SSR.full = SSR.full, SSR.diff = SSR.diff,
df1 = df1, df2 = df2, Fv = Fv, pvalue = pvalue)## $SSR.null
## [1] 820.425
##
## $SSR.full
## [1] 712.106
##
## $SSR.diff
## [1] 108.319
##
## $df1
## [1] 2
##
## $df2
## [1] 54
##
## $Fv
## [1] 4.106991
##
## $pvalue
## [1] 0.02186027Zapravo sve se radi preko lineranog modela i lm funkcije.
lm.1 <- lm(len ~ supp + dose, data = ToothGrowth)
lm.2 <- lm(len ~ supp * dose, data = ToothGrowth)
sum(residuals(lm.1)^2)## [1] 820.425sum(residuals(lm.2)^2)## [1] 712.106anova(lm.1,lm.2)## Analysis of Variance Table
##
## Model 1: len ~ supp + dose
## Model 2: len ~ supp * dose
## Res.Df RSS Df Sum of Sq F Pr(>F)
## 1 56 820.43
## 2 54 712.11 2 108.32 4.107 0.02186 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 14.6 Tukey test
TukeyHSD(a2_model)## Tukey multiple comparisons of means
## 95% family-wise confidence level
##
## Fit: aov(formula = len ~ supp + dose, data = ToothGrowth)
##
## $supp
## diff lwr upr p adj
## VC-OJ -3.7 -5.679762 -1.720238 0.0004293
##
## $dose
## diff lwr upr p adj
## 1-0.5 9.130 6.215909 12.044091 0e+00
## 2-0.5 15.495 12.580909 18.409091 0e+00
## 2-1 6.365 3.450909 9.279091 7e-06TukeyHSD(a2_inter)## Tukey multiple comparisons of means
## 95% family-wise confidence level
##
## Fit: aov(formula = len ~ supp * dose, data = ToothGrowth)
##
## $supp
## diff lwr upr p adj
## VC-OJ -3.7 -5.579828 -1.820172 0.0002312
##
## $dose
## diff lwr upr p adj
## 1-0.5 9.130 6.362488 11.897512 0.0e+00
## 2-0.5 15.495 12.727488 18.262512 0.0e+00
## 2-1 6.365 3.597488 9.132512 2.7e-06
##
## $`supp:dose`
## diff lwr upr p adj
## VC:0.5-OJ:0.5 -5.25 -10.048124 -0.4518762 0.0242521
## OJ:1-OJ:0.5 9.47 4.671876 14.2681238 0.0000046
## VC:1-OJ:0.5 3.54 -1.258124 8.3381238 0.2640208
## OJ:2-OJ:0.5 12.83 8.031876 17.6281238 0.0000000
## VC:2-OJ:0.5 12.91 8.111876 17.7081238 0.0000000
## OJ:1-VC:0.5 14.72 9.921876 19.5181238 0.0000000
## VC:1-VC:0.5 8.79 3.991876 13.5881238 0.0000210
## OJ:2-VC:0.5 18.08 13.281876 22.8781238 0.0000000
## VC:2-VC:0.5 18.16 13.361876 22.9581238 0.0000000
## VC:1-OJ:1 -5.93 -10.728124 -1.1318762 0.0073930
## OJ:2-OJ:1 3.36 -1.438124 8.1581238 0.3187361
## VC:2-OJ:1 3.44 -1.358124 8.2381238 0.2936430
## OJ:2-VC:1 9.29 4.491876 14.0881238 0.0000069
## VC:2-VC:1 9.37 4.571876 14.1681238 0.0000058
## VC:2-OJ:2 0.08 -4.718124 4.8781238 1.00000004.7 Nebalansirani dizajn
load("coffee.Rdata")
coffee %>% kable()| milk | sugar | babble |
|---|---|---|
| yes | real | 4.6 |
| no | fake | 4.4 |
| no | fake | 3.9 |
| yes | real | 5.6 |
| yes | real | 5.1 |
| no | real | 5.5 |
| yes | none | 3.9 |
| yes | none | 3.5 |
| yes | none | 3.7 |
| no | fake | 5.6 |
| no | fake | 4.7 |
| yes | fake | 5.9 |
| no | real | 6.0 |
| no | real | 5.4 |
| no | real | 6.6 |
| no | none | 5.8 |
| no | none | 5.3 |
| yes | fake | 5.7 |
coffee %>% count(milk, sugar)## milk sugar n
## 1 yes none 3
## 2 yes fake 2
## 3 yes real 3
## 4 no none 2
## 5 no fake 4
## 6 no real 44.7.1 Strategija tipa I
- Strategija tipa I gradi model postupno, počevši od najjednostavnijeg mogućeg modela i postupnog dodavanja novih varijabli.
- Nedostatak: ovisi o redoslijedu dodavanja novih varijabli u model.
model.1 <- lm(babble ~ sugar + milk + sugar:milk, coffee)
model.2 <- lm(babble ~ milk + sugar + sugar:milk, coffee)
anova(model.1)## Analysis of Variance Table
##
## Response: babble
## Df Sum Sq Mean Sq F value Pr(>F)
## sugar 2 3.5575 1.77876 6.7495 0.010863 *
## milk 1 0.9561 0.95611 3.6279 0.081061 .
## sugar:milk 2 5.9439 2.97193 11.2769 0.001754 **
## Residuals 12 3.1625 0.26354
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1anova(model.2)## Analysis of Variance Table
##
## Response: babble
## Df Sum Sq Mean Sq F value Pr(>F)
## milk 1 1.4440 1.44400 5.4792 0.037333 *
## sugar 2 3.0696 1.53482 5.8238 0.017075 *
## milk:sugar 2 5.9439 2.97193 11.2769 0.001754 **
## Residuals 12 3.1625 0.26354
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 14.7.2 Strategija tipa III
- Uvijek pokreće F-test s alternativnom hipotezom koja je jednaka punom modelu kojeg ispitujemo.
- Nulta hipoteza je jednaka modelu kojeg dobijemo tako da iz punog modela maknemo varijablu koju ispitujemo.
- Ovdje nemamo problem s redoslijedom varijabli koji postoji u strategiji tipa I.
- Međutim, ova strategija ovisi o načinu na koji se reprezentiraju faktorske varijable.
Za strategije tipa III i tipa II treba koristiti funkciju Anova iz car biblioteke.
Anova(model.1, type = 3)## Anova Table (Type III tests)
##
## Response: babble
## Sum Sq Df F value Pr(>F)
## (Intercept) 41.070 1 155.839 3.11e-08 ***
## sugar 5.880 2 11.156 0.001830 **
## milk 4.107 1 15.584 0.001936 **
## sugar:milk 5.944 2 11.277 0.001754 **
## Residuals 3.162 12
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Anova(model.2, type = 3)## Anova Table (Type III tests)
##
## Response: babble
## Sum Sq Df F value Pr(>F)
## (Intercept) 41.070 1 155.839 3.11e-08 ***
## milk 4.107 1 15.584 0.001936 **
## sugar 5.880 2 11.156 0.001830 **
## milk:sugar 5.944 2 11.277 0.001754 **
## Residuals 3.162 12
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Ukoliko promijenimo način reprezentacije faktorskih varijabli, rezultati se mogu značajno promijeniti.
model.H <- lm( babble ~ sugar * milk, coffee,
contrasts = list(milk = "contr.Helmert", sugar = "contr.Helmert"))
Anova(model.H, type = 3)## Anova Table (Type III tests)
##
## Response: babble
## Sum Sq Df F value Pr(>F)
## (Intercept) 434.29 1 1647.8882 3.231e-14 ***
## sugar 2.13 2 4.0446 0.045426 *
## milk 1.00 1 3.8102 0.074672 .
## sugar:milk 5.94 2 11.2769 0.001754 **
## Residuals 3.16 12
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 14.7.3 Strategija tipa II
- Uklanja probleme koji se javljau u prethodne dvije strategije pa je lakše interpretirati rezultate.
- Strategija je u potpunosti slična strategiji tipa III uz dodatni uvjet da se poštuje načelo marginalnosti.
- Načelo marginalnosti zabranjuje izostavljanje varijabli nižeg reda iz modela ako postoje varijable višeg reda koje ovise o njima.
Anova(model.1, type = 2)## Anova Table (Type II tests)
##
## Response: babble
## Sum Sq Df F value Pr(>F)
## sugar 3.0696 2 5.8238 0.017075 *
## milk 0.9561 1 3.6279 0.081061 .
## sugar:milk 5.9439 2 11.2769 0.001754 **
## Residuals 3.1625 12
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Anova(model.2, type = 2)## Anova Table (Type II tests)
##
## Response: babble
## Sum Sq Df F value Pr(>F)
## milk 0.9561 1 3.6279 0.081061 .
## sugar 3.0696 2 5.8238 0.017075 *
## milk:sugar 5.9439 2 11.2769 0.001754 **
## Residuals 3.1625 12
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Anova(model.H, type = 2)## Anova Table (Type II tests)
##
## Response: babble
## Sum Sq Df F value Pr(>F)
## sugar 3.0696 2 5.8238 0.017075 *
## milk 0.9561 1 3.6279 0.081061 .
## sugar:milk 5.9439 2 11.2769 0.001754 **
## Residuals 3.1625 12
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1ovdje je opis u fusnoti↩︎